Overview
The Antimicrobial Peptide (AMP) Scanner is a free service to predict if a protein sequence may be an AMP active against Gram-positive and/or Gram-negative bacteria (but not fungi and viruses). Version 2 of the server uses a deep neural network (DNN) to classify 1-50,000 query peptides submitted in valid FASTA format as "AMPs" or "Non-AMPs." Specifically, the algorithm considers a peptide with a prediction probability > 0.5 as an AMP.
Prediction results can be downloaded as a zip file and will be available online for 72 hours before being deleted. When submitting over 2,000 sequences, we recommend using the 'file upload' option rather than pasting sequences into the text box. If you need to submit many large batches of sequences, please get in touch with us first so we can assist you.
As the network was trained using proteins between 10-200 amino acids (AA) in length, we highly recommend submitting queries in this range using the standard 20AA for best results. Predictions for queries less than 10AA will be returned with a warning. For longer sequences, only the last 200AA will be considered. For sequences longer than 200AA, we recommend using "Proteome Scan Mode" in AMP Scanner Ver. 1 to identify promising segments, and then submitting those results to Ver. 2 of the program. Please note, prediction probabilities relate only to AMP vs Non-AMP activity. In other words, probabilities for two sequences predicted as AMPs should not be compared and interpreted as one having "better" or "worse" activity against any given bacteria.
DNN Structure
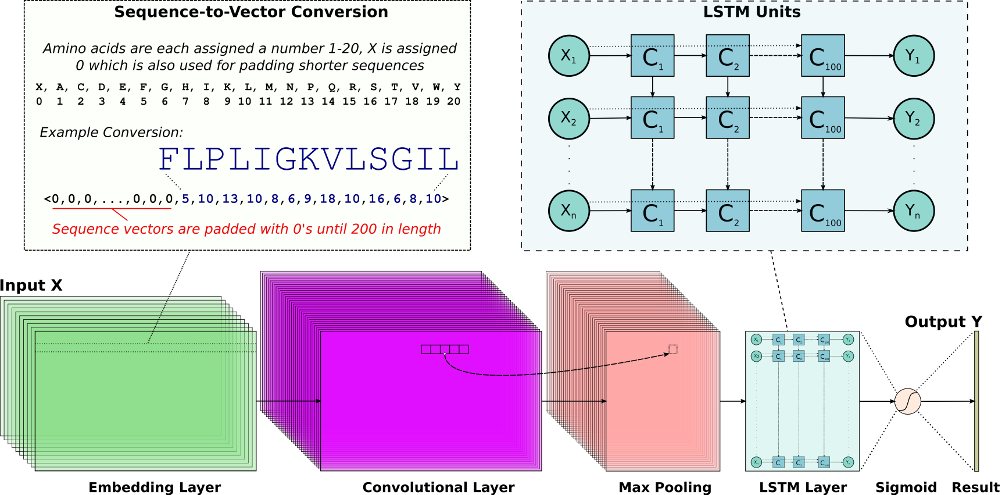
The DNN uses a mix of convolutional (Conv) and recurrent long short-term memory (LSTM) layers as outlined above. Peptide sequences are first encoded into uniform numerical vectors of length 200. Shorter sequences padded with 0's in front which the network ignores. These vectors (X) are fed to an embedding layer of length 128, followed by a Conv layer comprised of 64 filters. Each of these filters undergoes a 1D convolution and is down-sampled via a maximal pooling layer of size 5. Next, an LSTM layer with 100 units allows the DNN to remember or ignore old information passed along the horizontal dotted arrows extending from each Xi input. The final output from the DNN is passed through a sigmoid function, so that predictions (Y) are scaled between 0 and 1. Our network was trained using the Keras framework using a Tensorflow back-end. For additional details, please see the paper.
Data Sets
Data sets and models are occasionally updated as more AMP sequences become available and are described in the "News" section when they are released. The "Original Production" model described in the paper is trained on 1,778 AMPs from the APD database (Vr.3) listed as having activity against Gram-positive and/or Gram-negative bacteria and 1,778 non-AMPs randomly selected from Uniprot and has a classification accuracy of 98.96% on these sequences as detailed in the paper. The "Original Training" model from the paper is trained using about 60% of the data set, has a 91.01% classification accuracy on the remaining testing data, and can be used for those interested in comparing prediction methods.
FASTA files for the AMP and Non-AMPs used to train, validate, and test the DNN models in the paper can be downloaded here (.zip 91.9kB)
Code and Model Files
Code and pretrained model files are available on GitHub and as a Docker image for those interested in making offline predictions or training their own models. For details, please see the GitHub repository here: https://github.com/dan-veltri/amp-scanner-v2. If you encounter problems, please submit an issue on GitHub or contact us.
